
If your Cytoscape session starts acting funny, go to Help > Error Console. A global minimum size is set for all panels so that users can manually resize no matter which panel they are viewing. We revamped the behavior of CytoPanels so that each plugins preferred panel size will be automatically fit when the user switches to them. We've overhauled the user interfaces controlling the mapping of continuous values to visual styles, like Node Size and Node Color, to provide more intuitive and consistent controls. Minimum System Requirements Java 1.5+ JVM (1.6 is required for Cytoscape 2.8+)Note: Queries that use more than 1.5 GB of RAM require Cytoscape to be launched with a 64-bit JVM Cytoscape2.6.3+, 2.7+, or 2.8+ (does not work with 3.0 yet) 2 GB RAM Installation Instructions Adjust the amount of memory allocated to Cytoscapeto at least 1. The new process avoids system crashes that were occurring with 2.8.1. The process attempts to optimize memory settings based on your hardware. We have improved the memory detection and allocation process that occurs when Cytoscape is first installed and launched. We hope the changes will make the Cytoscape user experience more pleasant and consistent.
#Cytoscape 2.8 full
This means thatĬytoscape now has a full suite of network analysis and graphing routines available right Provided their Network Analyzer plugin to Cytoscape as a core plugin. Mario Albrecht and his team at the Max-Planck Institute for Informatics have graciously Equations are not meant for heavyĭata analysis, but rather as a simple way to manipulate attribute values within Cytoscape.
#Cytoscape 2.8 how to
How to properly specify the arguments to a command. We've providedĪn equation editor to help users understand the functions that are available as well as Of Excel-like functions which dynamically compute values as attributes.
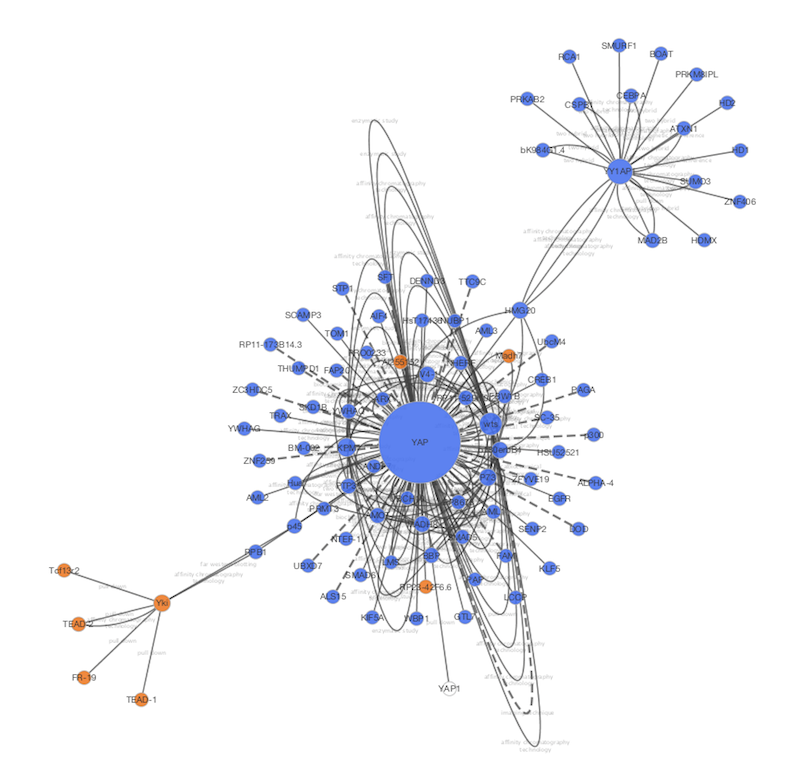
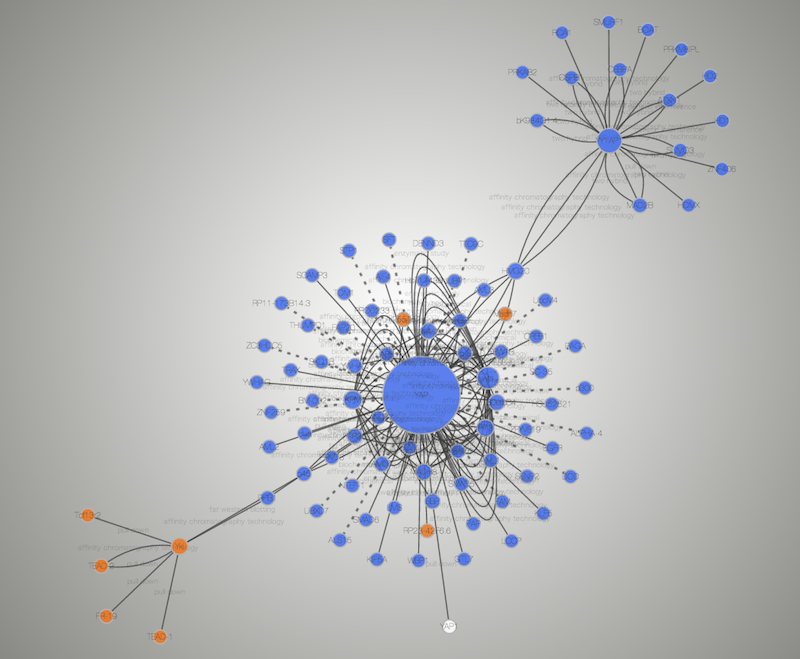
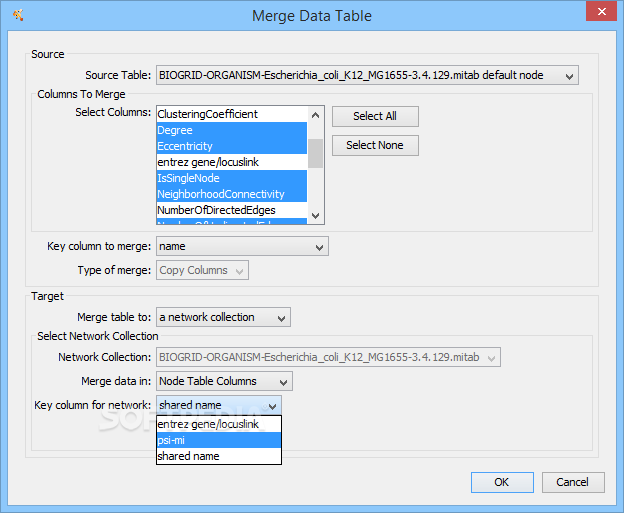
We've had many requests over the years to provide spreadsheet like functionality in theĪttribute browser, and now we have! Cells in the attribute browser now accept a subset Users can control both the size and placement of Images to be mapped independently onto nodes. Version 2. To connect these images to nodes, the VizMapper provides visual properties that allow up to nine Cytoscape is a popular bioinformatics package for biological network visualization and data integration. May also use the C圜ustomGraphics interface to create images dynamically within plugins. ImagesĬan be loaded manually with our image browser or specified as URLs in attributes. We've added the ability to associate images and other custom graphics with nodes. degree or clustering index).New Features in Cytoscape 2.8.0 Custom Graphics
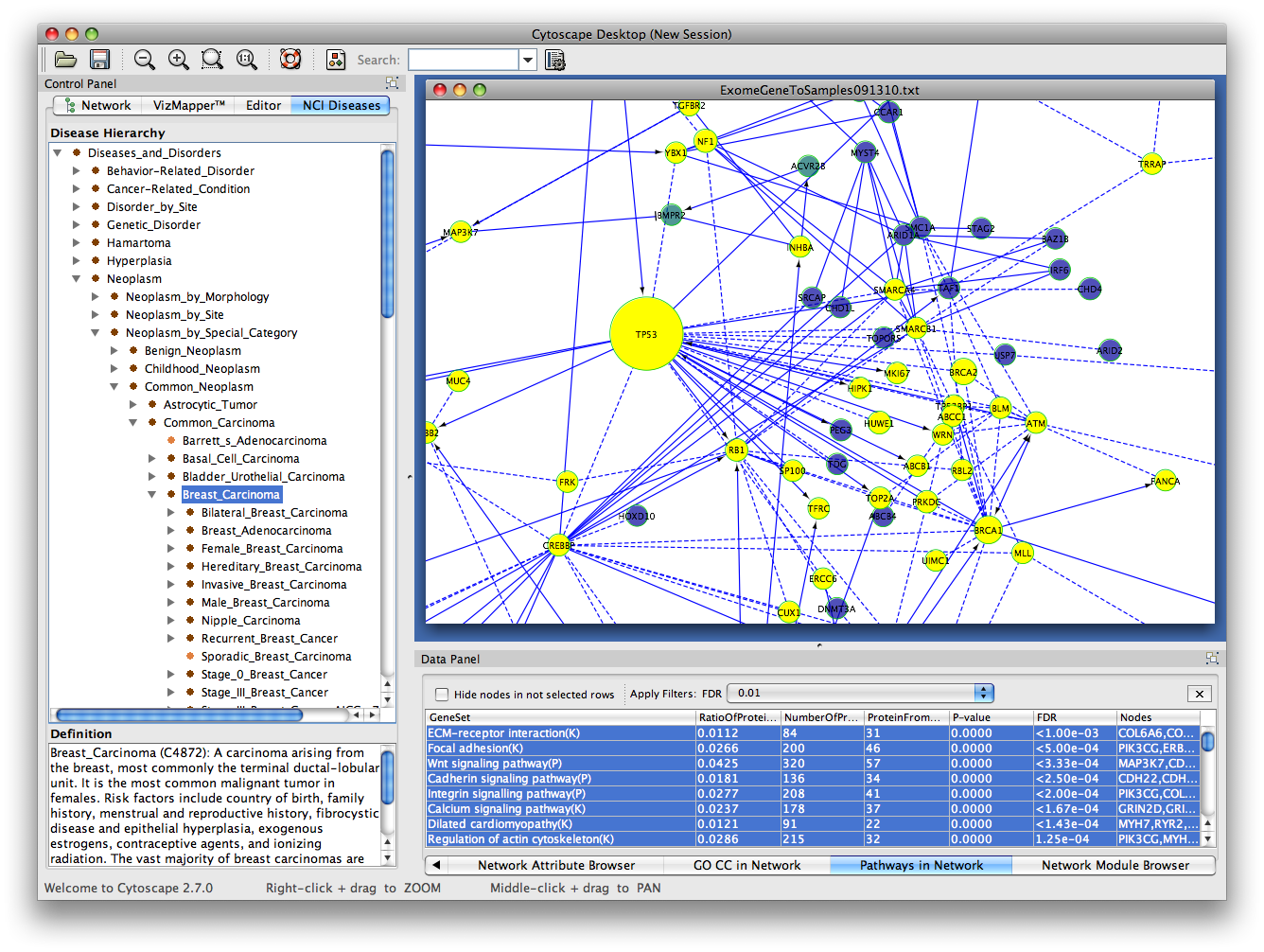
Finally, to enrich the visual exploration, it is possible to visually render local topological properties of the network (e.g. The plugin also allows the extraction of parts of the network that contain a selected subset of reactions. the organism or perturbation under study), we propose facilities to edit/select putative biochemical transformations. Since the definition of this list is closely related to experimentation (i.e. Inference requires a list of potential biochemical transformations. Here we present a new plugin for Cytoscape dedicated to the inference and visualization of high-resolution metabolomic networks.
#Cytoscape 2.8 software
There is currently no available software that allows inference and visualization of such high-resolution metabolomic networks directly from raw data. To analyze, explore and interpret these two kinds of relations, powerful visualization tools are required. The combination of these two inference methods generates networks containing hundreds of nodes (metabolites) and hundreds of predicted edges (biochemical reactions and/or high correlations). Moreover, perturbation studies allow the use of correlation analysis to infer/confirm links between metabolites that correlate across various conditions. Such high-resolution data has also been used to predict ab initio biochemical interactions between metabolites. Recently, ultra high-resolution mass spectrometry (FTICR-MS or Orbitrap) has been successfully used in metabolomic studies. Various spectrometric technologies are capable of identifying thousands of metabolites. Metabolomics aims at the identification and quantification of all metabolites that are present in a cell, tissue or biofluid at a given moment and under particular conditions.


 0 kommentar(er)
0 kommentar(er)
